StagewiseNN - Building developmental tree from single-cell data¶
StagewiseNN is a computational tool for constructing developmental (lineage) tree from multi-staged single-cell RNA-seq data.
It starts from building a single-cell graph by connecting each cell to its k-nearest neighbors in the parent stage, followed by the voting-based tree-construction and an adaptive cluster refinement.
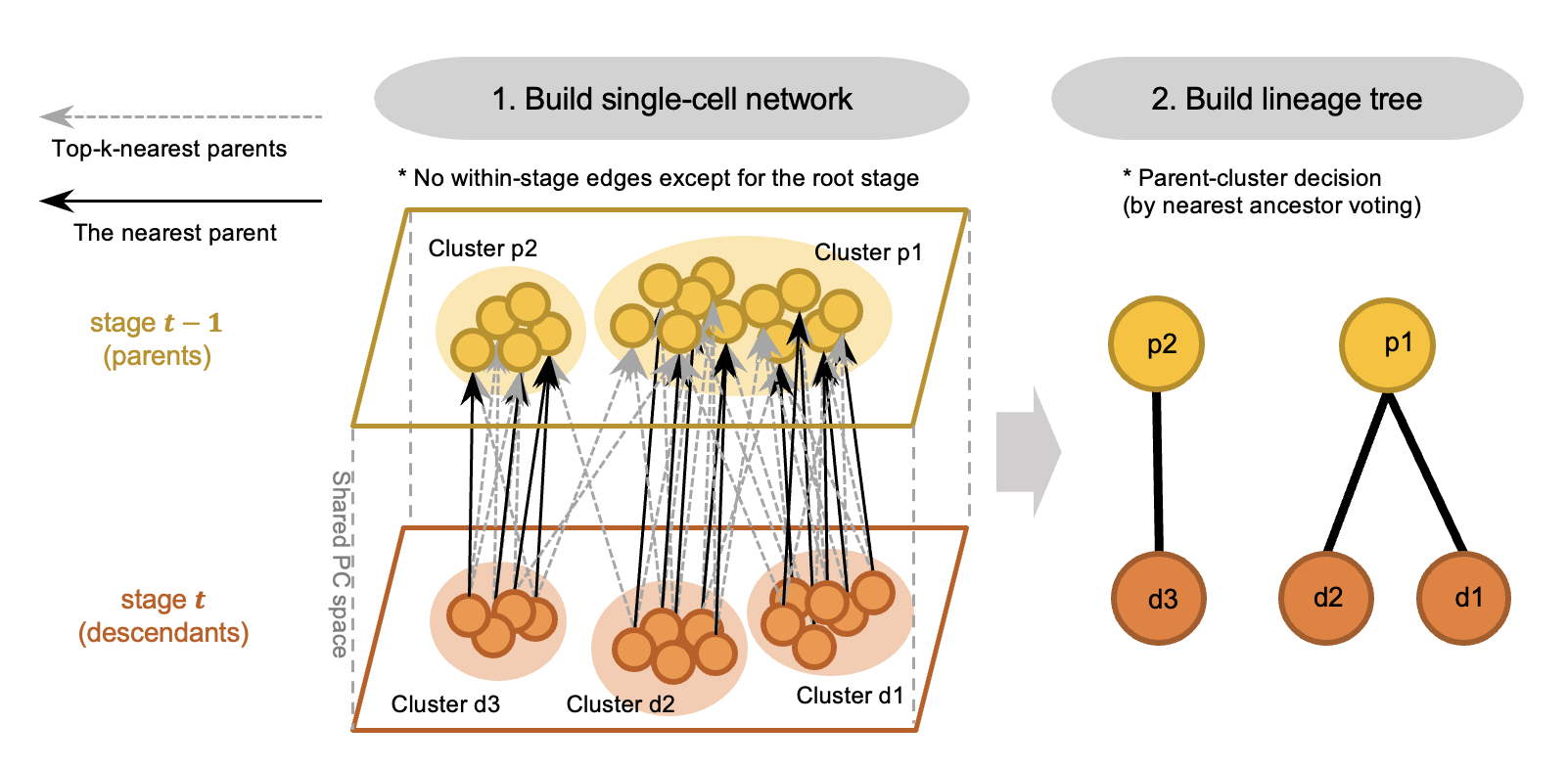
The single-cell graph can be further visualized using graph embedding methods, e.g. UMAP, SPRING.
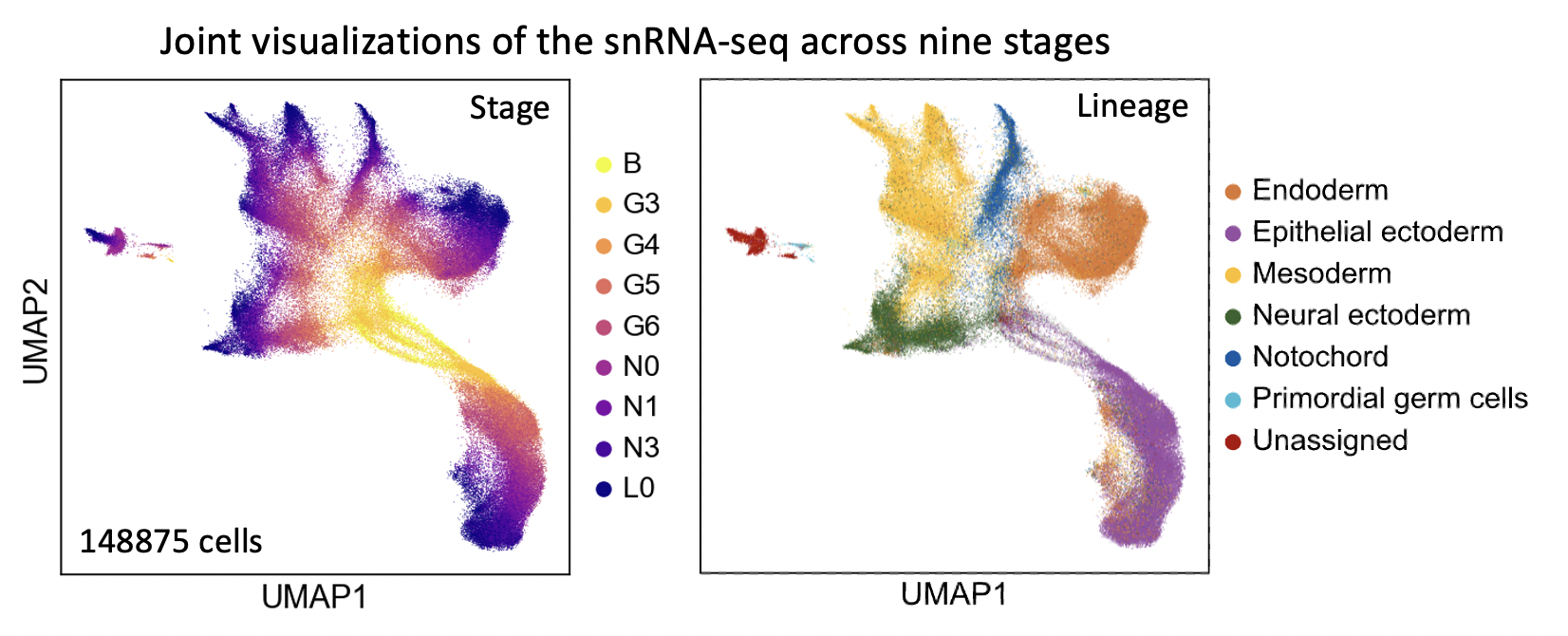
We have used it to build the developmental tree from the snRNA-seq of amphioxus embryonic cells, across nine developmental stages (“B”, “G3”, “G4”, “G5”, “G6”, “N0”, “N1”, “N3”, “L0”), where seven major lineages were recognized.
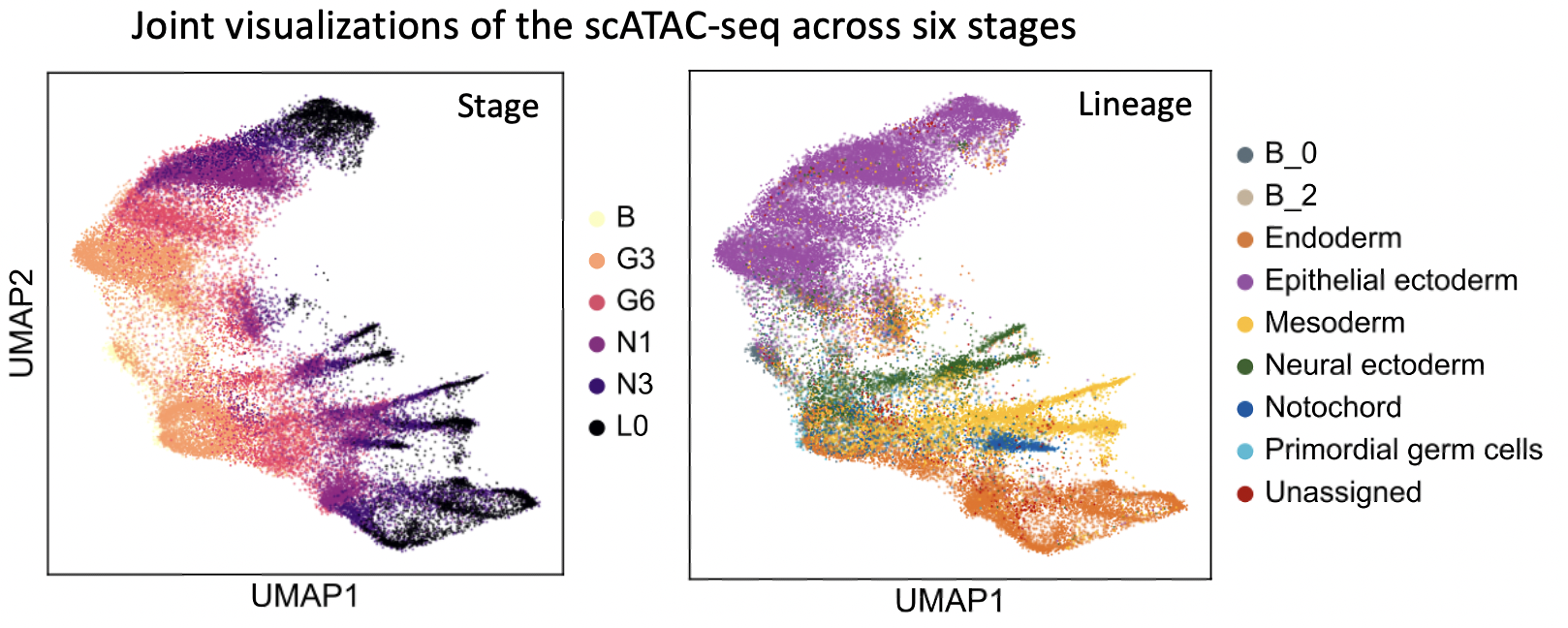
StagewiseNN can also be applied on scATAC-seq data sampled at multiple timepoints, once the peak-by-cell matrix is transformed into a gene-by-cell matrix (i.e., the gene activities).
Installation¶
Requirements:
python >= 3.6
scanpy: https://scanpy.readthedocs.io/en/stable/installation.html
scikit-learn: https://pypi.org/project/scikit-learn/
Install stagewiseNN with PyPI, run:
pip install swnn
Alternatively, install from source code:
git clone https://github.com/zhanglabtools/stagewiseNN.git
cd stagewiseNN
python setup.py install
Usage¶
See Tutorial for using stagewiseNN for detailed guide.
import swnn
# ====== Inputs ======
# data_matrix = ..
# stage_labels = ..
# group_labels = ..
# stage_order = [f'stage_{i}' for i in range(5)]
builder = swnn.Builder(stage_order=stage_order)
# step1: building a (stage-preserved) single-cell graph
distmat, connect = builder.build_graph(
X=data_matrix, stage_lbs=stage_labels,
)
# step2: build a developmental tree from the single-cell graph
edgedf, refined_group_lbs = builder.build_tree(group_labels, stage_labels,)
Contribute¶
Issue Tracker: https://github.com/XingyanLiu/stagewiseNN/issues
Source Code:
https://github.com/XingyanLiu/stagewiseNN (the developmental version)